Our pipeline
ROME is designing therapies with the potential to modulate disease at the source. We are pursuing drug discovery and development by targeting three disease-relevant consequences of repeat dysregulation.
Viral mimicry
We are developing novel therapeutics that block repeat-induced proinflammatory responses.
Tumor antigens
We are developing novel therapeutics that specifically target cancer cells expressing abnormally high levels of repeat-derived antigens.

Genomic instability
We are developing novel therapeutics targeting repeat-induced genomic instability associated with disease.
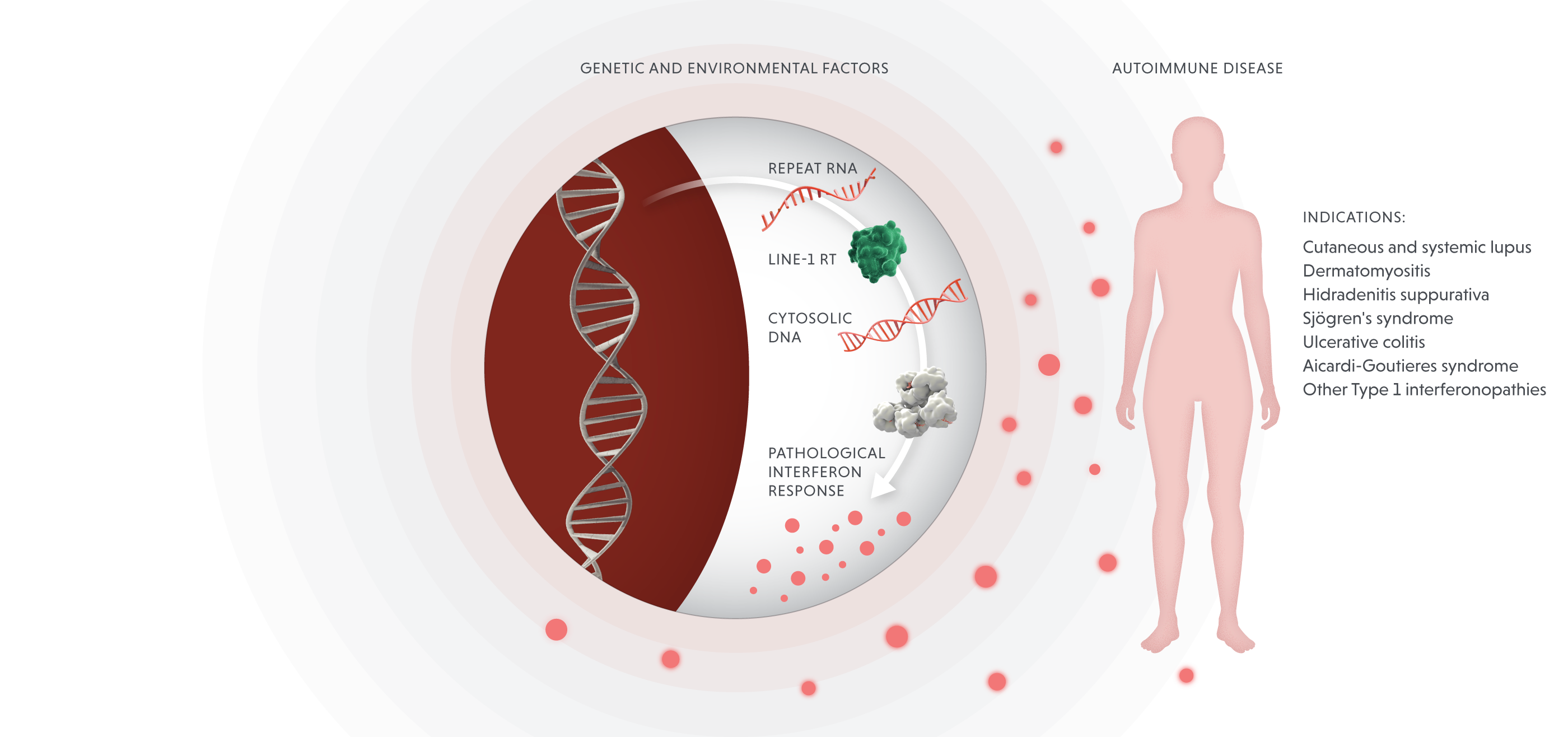
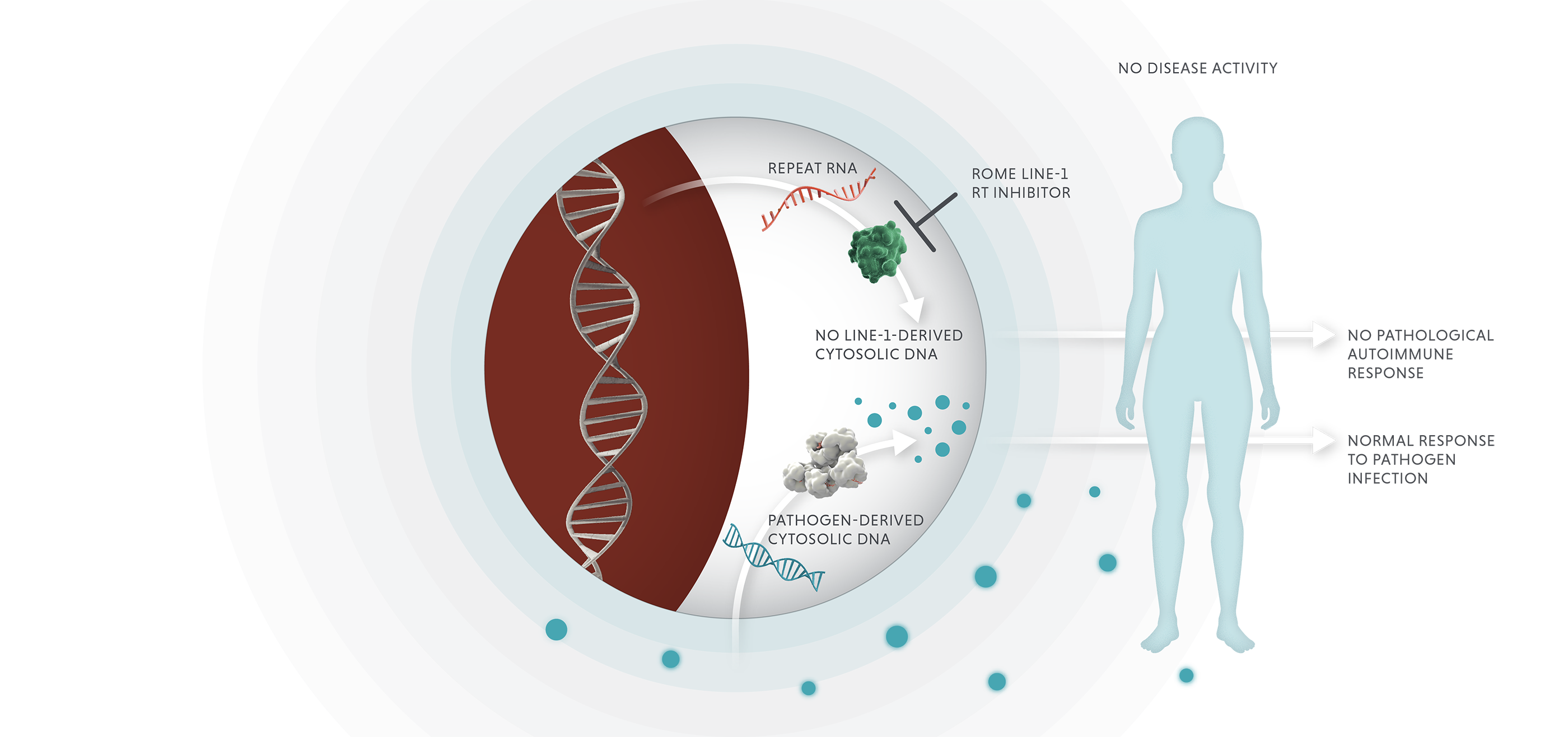
Blocking LINE-1 reverse transcriptase in autoimmune disease
Our data science platform
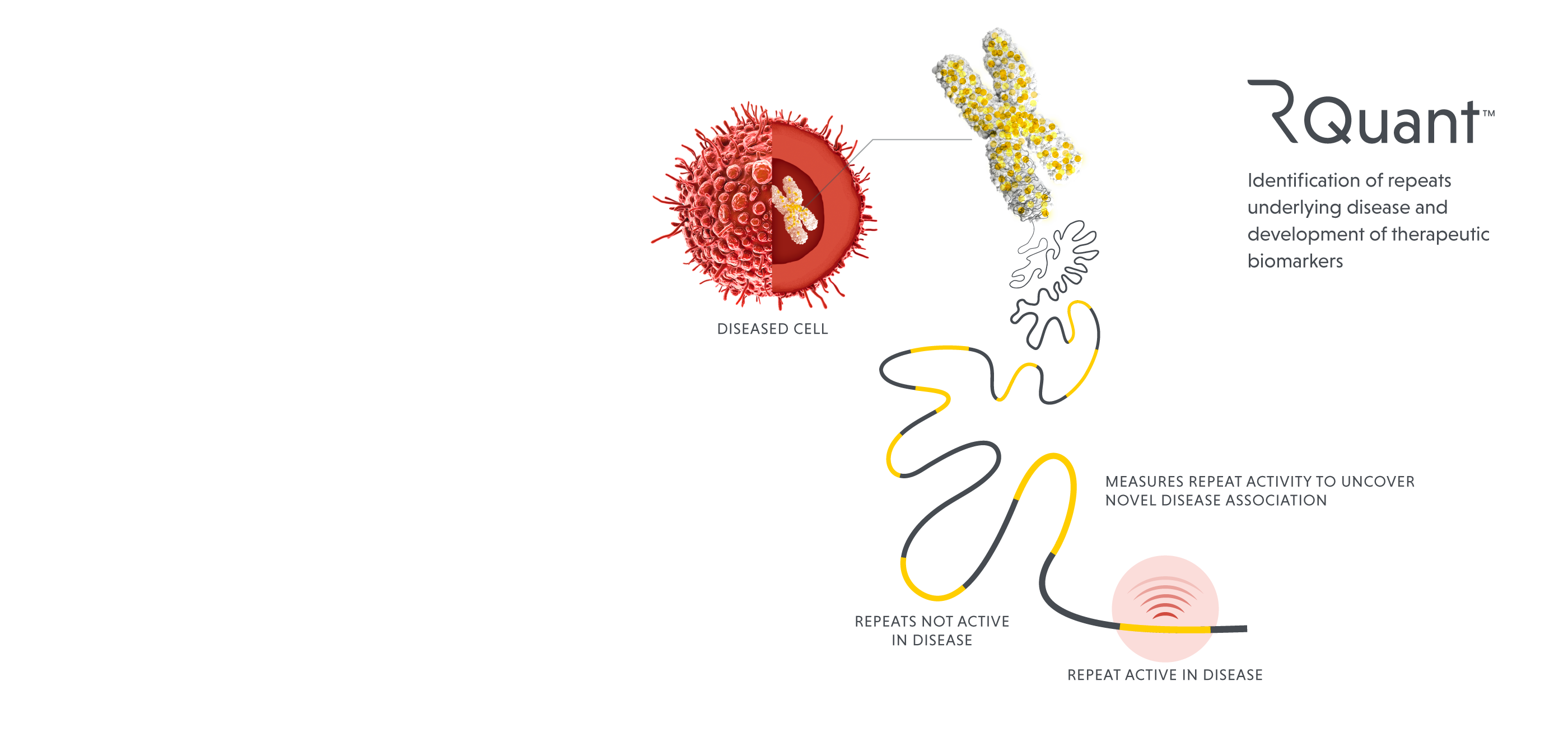
Our data science platform is designed to illuminate the target-rich yet underexplored dark genome, using proprietary tools and algorithms to identify and advance new and better therapies. High-resolution mapping and annotation techniques first identify functionally active repeats, then machine learning methods quantify their expression to determine which of the mapped repeats are driving disease. The platform has shown demonstrated capabilities in target discovery and indication selection; it can also inform clinical trial design, biomarker discovery and potentially provide a means to measure an individual patient’s response to therapies.
ROME’s data science platform is the first and most comprehensive set of unified genomic analysis tools to specifically probe the repetitive elements and their function. Using conventional approaches, it is easy to inaccurately estimate the abundance of repeat-related activities since the sequences of functional and nonfunctional repeat elements are often very similar to each other. Our platform has been tuned to focus on the individual repeats that are active, yielding insights about activities within the cell that drive disease.
Partnering
ROME has deep expertise in the biology of genomic repeats and specialized repeat-oriented data science tools with a variety of applications. We are open to and interested in applying these capabilities in new settings through strategic collaboration.
CONTACT: bd@rometx.com